25. A First Look at the Kalman Filter#
Contents
In addition to what’s in Anaconda, this lecture will need the following libraries:
!pip install quantecon
Show code cell output
Requirement already satisfied: quantecon in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (0.8.1)
Requirement already satisfied: numba>=0.49.0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from quantecon) (0.60.0)
Requirement already satisfied: numpy>=1.17.0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from quantecon) (1.26.4)
Requirement already satisfied: requests in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from quantecon) (2.32.3)
Requirement already satisfied: scipy>=1.5.0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from quantecon) (1.13.1)
Requirement already satisfied: sympy in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from quantecon) (1.14.0)
Requirement already satisfied: llvmlite<0.44,>=0.43.0dev0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from numba>=0.49.0->quantecon) (0.43.0)
Requirement already satisfied: charset-normalizer<4,>=2 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from requests->quantecon) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from requests->quantecon) (3.7)
Requirement already satisfied: urllib3<3,>=1.21.1 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from requests->quantecon) (2.2.3)
Requirement already satisfied: certifi>=2017.4.17 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from requests->quantecon) (2024.8.30)
Requirement already satisfied: mpmath<1.4,>=1.1.0 in /home/runner/miniconda3/envs/quantecon/lib/python3.12/site-packages (from sympy->quantecon) (1.3.0)
25.1. Overview#
This lecture provides a simple and intuitive introduction to the Kalman filter, for those who either
have heard of the Kalman filter but don’t know how it works, or
know the Kalman filter equations, but don’t know where they come from
For additional (more advanced) reading on the Kalman filter, see
[Ljungqvist and Sargent, 2018], section 2.7
The second reference presents a comprehensive treatment of the Kalman filter.
Required knowledge: Familiarity with matrix manipulations, multivariate normal distributions, covariance matrices, etc.
We’ll need the following imports:
import matplotlib.pyplot as plt
from scipy import linalg
import numpy as np
import matplotlib.cm as cm
from quantecon import Kalman, LinearStateSpace
from scipy.stats import norm
from scipy.integrate import quad
from scipy.linalg import eigvals
25.2. The Basic Idea#
The Kalman filter has many applications in economics, but for now let’s pretend that we are rocket scientists.
A missile has been launched from country Y and our mission is to track it.
Let
At the present moment in time, the precise location
One way to summarize our knowledge is a point prediction
But what if the President wants to know the probability that the missile is currently over the Sea of Japan?
Then it is better to summarize our initial beliefs with a bivariate probability density
The density
To keep things tractable in our example, we assume that our prior is Gaussian.
In particular, we take
where
This density
# Set up the Gaussian prior density p
Σ = [[0.4, 0.3], [0.3, 0.45]]
Σ = np.matrix(Σ)
x_hat = np.matrix([0.2, -0.2]).T
# Define the matrices G and R from the equation y = G x + N(0, R)
G = [[1, 0], [0, 1]]
G = np.matrix(G)
R = 0.5 * Σ
# The matrices A and Q
A = [[1.2, 0], [0, -0.2]]
A = np.matrix(A)
Q = 0.3 * Σ
# The observed value of y
y = np.matrix([2.3, -1.9]).T
# Set up grid for plotting
x_grid = np.linspace(-1.5, 2.9, 100)
y_grid = np.linspace(-3.1, 1.7, 100)
X, Y = np.meshgrid(x_grid, y_grid)
def bivariate_normal(x, y, σ_x=1.0, σ_y=1.0, μ_x=0.0, μ_y=0.0, σ_xy=0.0):
"""
Compute and return the probability density function of bivariate normal
distribution of normal random variables x and y
Parameters
----------
x : array_like(float)
Random variable
y : array_like(float)
Random variable
σ_x : array_like(float)
Standard deviation of random variable x
σ_y : array_like(float)
Standard deviation of random variable y
μ_x : scalar(float)
Mean value of random variable x
μ_y : scalar(float)
Mean value of random variable y
σ_xy : array_like(float)
Covariance of random variables x and y
"""
x_μ = x - μ_x
y_μ = y - μ_y
ρ = σ_xy / (σ_x * σ_y)
z = x_μ**2 / σ_x**2 + y_μ**2 / σ_y**2 - 2 * ρ * x_μ * y_μ / (σ_x * σ_y)
denom = 2 * np.pi * σ_x * σ_y * np.sqrt(1 - ρ**2)
return np.exp(-z / (2 * (1 - ρ**2))) / denom
def gen_gaussian_plot_vals(μ, C):
"Z values for plotting the bivariate Gaussian N(μ, C)"
m_x, m_y = float(μ[0,0]), float(μ[1,0])
s_x, s_y = np.sqrt(C[0, 0]), np.sqrt(C[1, 1])
s_xy = C[0, 1]
return bivariate_normal(X, Y, s_x, s_y, m_x, m_y, s_xy)
# Plot the figure
fig, ax = plt.subplots(figsize=(10, 8))
ax.grid()
Z = gen_gaussian_plot_vals(x_hat, Σ)
ax.contourf(X, Y, Z, 6, alpha=0.6, cmap=cm.jet)
cs = ax.contour(X, Y, Z, 6, colors="black")
ax.clabel(cs, inline=1, fontsize=10)
plt.show()
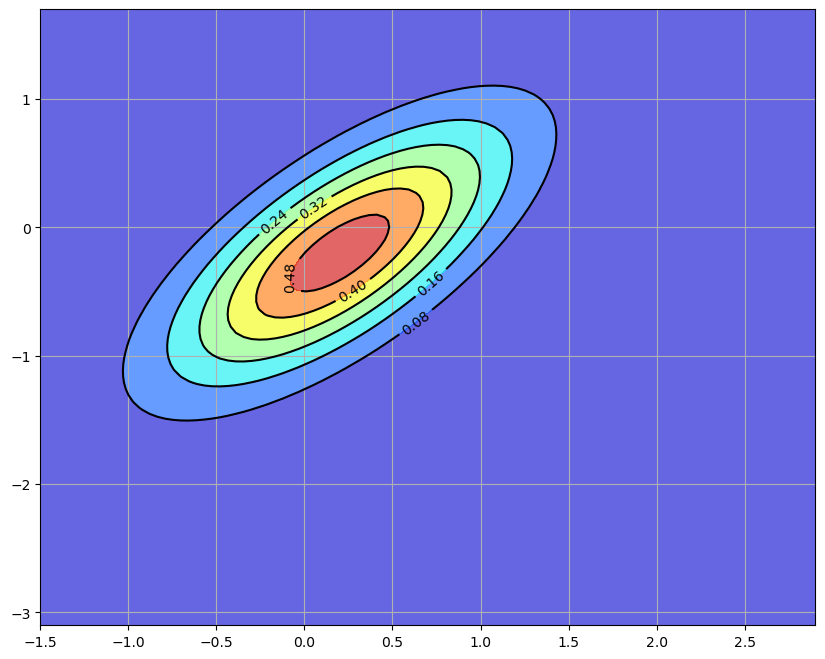
25.2.1. The Filtering Step#
We are now presented with some good news and some bad news.
The good news is that the missile has been located by our sensors, which report that the current location is
The next figure shows the original prior
fig, ax = plt.subplots(figsize=(10, 8))
ax.grid()
Z = gen_gaussian_plot_vals(x_hat, Σ)
ax.contourf(X, Y, Z, 6, alpha=0.6, cmap=cm.jet)
cs = ax.contour(X, Y, Z, 6, colors="black")
ax.clabel(cs, inline=1, fontsize=10)
ax.text(float(y[0].item()), float(y[1].item()), "$y$", fontsize=20, color="black")
plt.show()
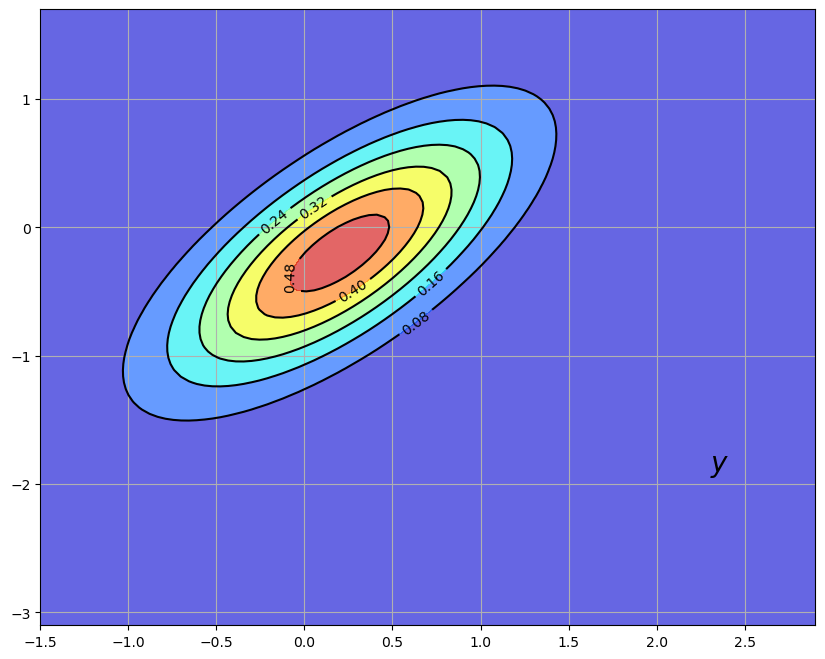
The bad news is that our sensors are imprecise.
In particular, we should interpret the output of our sensor not as
Here
How then should we combine our prior
As you may have guessed, the answer is to use Bayes’ theorem, which tells
us to update our prior
where
In solving for
In view of (25.3), the conditional density
Because we are in a linear and Gaussian framework, the updated density can be computed by calculating population linear regressions.
In particular, the solution is known 1 to be
where
Here
This new density
The original density is left in as contour lines for comparison
fig, ax = plt.subplots(figsize=(10, 8))
ax.grid()
Z = gen_gaussian_plot_vals(x_hat, Σ)
cs1 = ax.contour(X, Y, Z, 6, colors="black")
ax.clabel(cs1, inline=1, fontsize=10)
M = Σ * G.T * linalg.inv(G * Σ * G.T + R)
x_hat_F = x_hat + M * (y - G * x_hat)
Σ_F = Σ - M * G * Σ
new_Z = gen_gaussian_plot_vals(x_hat_F, Σ_F)
cs2 = ax.contour(X, Y, new_Z, 6, colors="black")
ax.clabel(cs2, inline=1, fontsize=10)
ax.contourf(X, Y, new_Z, 6, alpha=0.6, cmap=cm.jet)
ax.text(float(y[0].item()), float(y[1].item()), "$y$", fontsize=20, color="black")
plt.show()
Our new density twists the prior
In generating the figure, we set
25.2.2. The Forecast Step#
What have we achieved so far?
We have obtained probabilities for the current location of the state (missile) given prior and current information.
This is called “filtering” rather than forecasting because we are filtering out noise rather than looking into the future.
But now let’s suppose that we are given another task: to predict the location of the missile after one unit of time (whatever that may be) has elapsed.
To do this we need a model of how the state evolves.
Let’s suppose that we have one, and that it’s linear and Gaussian. In particular,
Our aim is to combine this law of motion and our current distribution
In view of (25.5), all we have to do is introduce a random vector
Since linear combinations of Gaussians are Gaussian,
Elementary calculations and the expressions in (25.4) tell us that
and
The matrix
The subscript
Using this notation, we can summarize our results as follows.
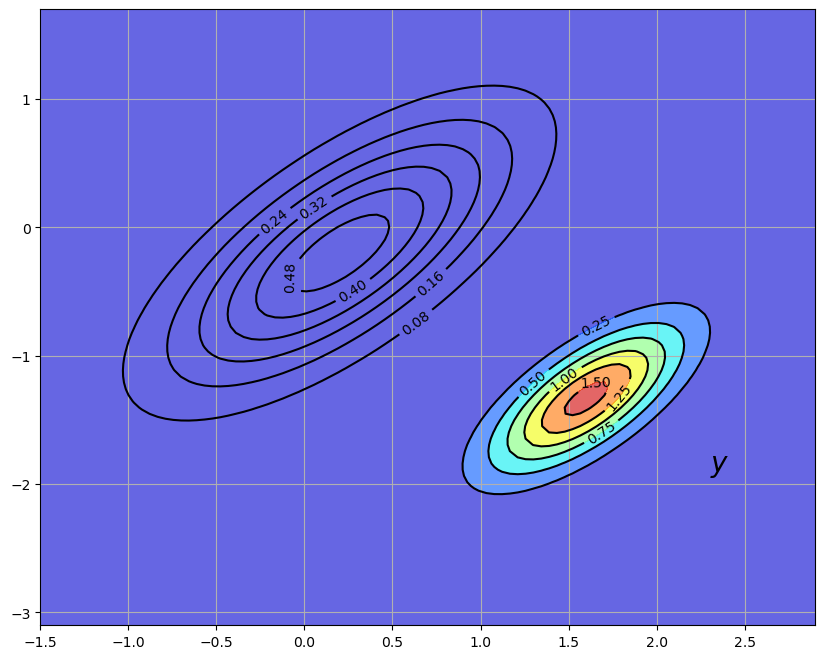
Our updated prediction is the density
The density
The predictive distribution is the new density shown in the following figure, where the update has used parameters.
fig, ax = plt.subplots(figsize=(10, 8))
ax.grid()
# Density 1
Z = gen_gaussian_plot_vals(x_hat, Σ)
cs1 = ax.contour(X, Y, Z, 6, colors="black")
ax.clabel(cs1, inline=1, fontsize=10)
# Density 2
M = Σ * G.T * linalg.inv(G * Σ * G.T + R)
x_hat_F = x_hat + M * (y - G * x_hat)
Σ_F = Σ - M * G * Σ
Z_F = gen_gaussian_plot_vals(x_hat_F, Σ_F)
cs2 = ax.contour(X, Y, Z_F, 6, colors="black")
ax.clabel(cs2, inline=1, fontsize=10)
# Density 3
new_x_hat = A * x_hat_F
new_Σ = A * Σ_F * A.T + Q
new_Z = gen_gaussian_plot_vals(new_x_hat, new_Σ)
cs3 = ax.contour(X, Y, new_Z, 6, colors="black")
ax.clabel(cs3, inline=1, fontsize=10)
ax.contourf(X, Y, new_Z, 6, alpha=0.6, cmap=cm.jet)
ax.text(float(y[0].item()), float(y[1].item()), "$y$", fontsize=20, color="black")
plt.show()
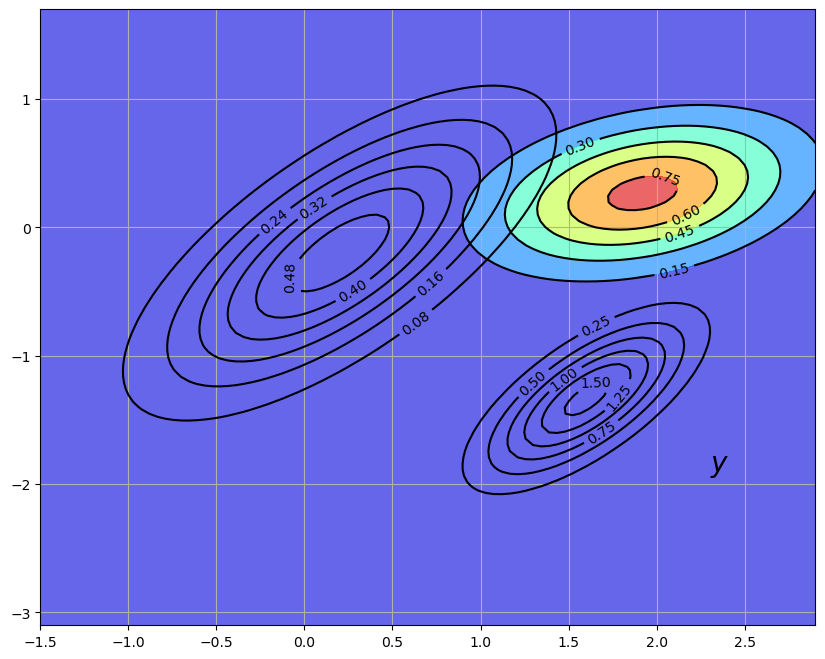
25.2.3. The Recursive Procedure#
Let’s look back at what we’ve done.
We started the current period with a prior
We then used the current measurement
Finally, we used the law of motion (25.5) for
If we now step into the next period, we are ready to go round again, taking
Swapping notation
Start the current period with prior
Observe current measurement
Compute the filtering distribution
Compute the predictive distribution
Increment
Repeating (25.6), the dynamics for
These are the standard dynamic equations for the Kalman filter (see, for example, [Ljungqvist and Sargent, 2018], page 58).
25.3. Convergence#
The matrix
Apart from special cases, this uncertainty will never be fully resolved, regardless of how much time elapses.
One reason is that our prediction
Even if we know the precise value of
Since the shock
However, it is certainly possible that
To study this topic, let’s expand the second equation in (25.7):
This is a nonlinear difference equation in
A fixed point of (25.8) is a constant matrix
Equation (25.8) is known as a discrete-time Riccati difference equation.
Equation (25.9) is known as a discrete-time algebraic Riccati equation.
Conditions under which a fixed point exists and the sequence
A sufficient (but not necessary) condition is that all the eigenvalues
(This strong condition assures that the unconditional distribution of
In this case, for any initial choice of
25.4. Implementation#
The class Kalman from the QuantEcon.py package implements the Kalman filter
Instance data consists of:
the moments
An instance of the LinearStateSpace class from QuantEcon.py.
The latter represents a linear state space model of the form
where the shocks
To connect this with the notation of this lecture we set
The class
Kalmanfrom the QuantEcon.py package has a number of methods, some that we will wait to use until we study more advanced applications in subsequent lectures.Methods pertinent for this lecture are:
prior_to_filtered, which updatesfiltered_to_forecast, which updates the filtering distribution to the predictive distribution – which becomes the new priorupdate, which combines the last two methodsa
stationary_values, which computes the solution to (25.9) and the corresponding (stationary) Kalman gain
You can view the program on GitHub.
25.5. Exercises#
Exercise 25.1
Consider the following simple application of the Kalman filter, loosely based on [Ljungqvist and Sargent, 2018], section 2.9.2.
Suppose that
all variables are scalars
the hidden state
State dynamics are therefore given by (25.5) with
The measurement equation is
The task of this exercise to simulate the model and, using the code from kalman.py, plot the first five predictive densities
As shown in [Ljungqvist and Sargent, 2018], sections 2.9.1–2.9.2, these distributions asymptotically put all mass on the unknown value
In the simulation, take
Your figure should – modulo randomness – look something like this
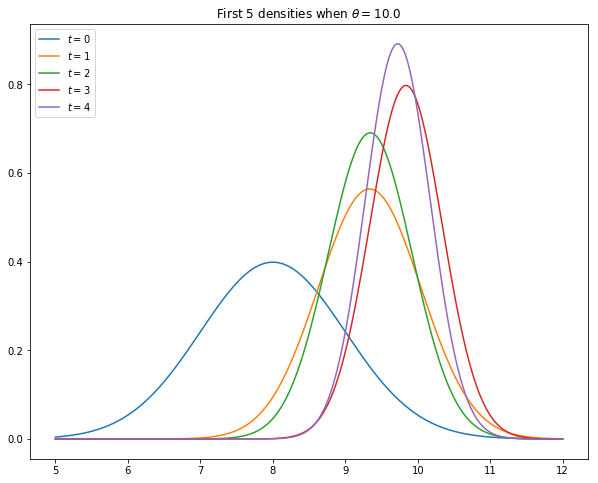
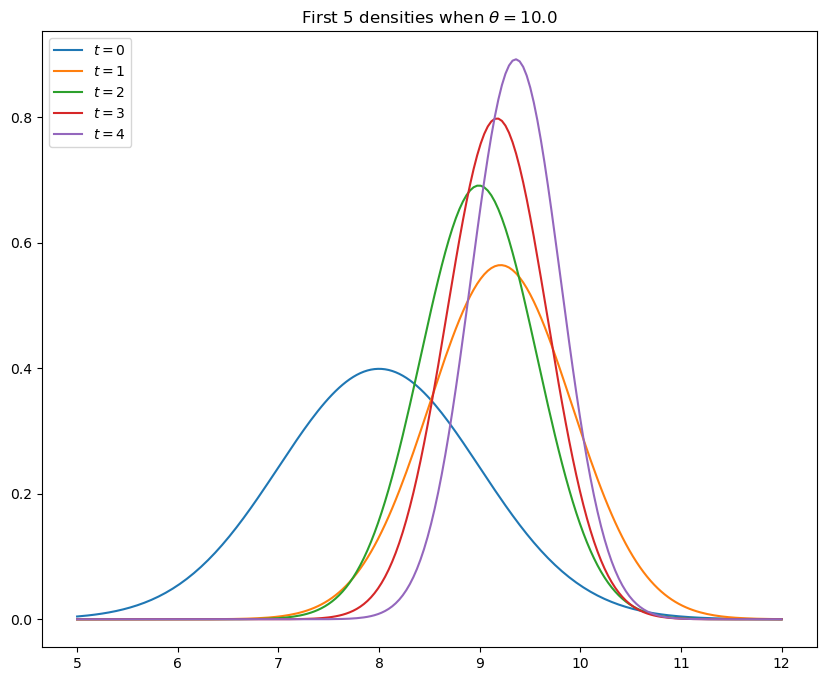
Solution to Exercise 25.1
# Parameters
θ = 10 # Constant value of state x_t
A, C, G, H = 1, 0, 1, 1
ss = LinearStateSpace(A, C, G, H, mu_0=θ)
# Set prior, initialize kalman filter
x_hat_0, Σ_0 = 8, 1
kalman = Kalman(ss, x_hat_0, Σ_0)
# Draw observations of y from state space model
N = 5
x, y = ss.simulate(N)
y = y.flatten()
# Set up plot
fig, ax = plt.subplots(figsize=(10,8))
xgrid = np.linspace(θ - 5, θ + 2, 200)
for i in range(N):
# Record the current predicted mean and variance
m, v = [float(z) for z in (kalman.x_hat.item(), kalman.Sigma.item())]
# Plot, update filter
ax.plot(xgrid, norm.pdf(xgrid, loc=m, scale=np.sqrt(v)), label=f'$t={i}$')
kalman.update(y[i])
ax.set_title(f'First {N} densities when $\\theta = {θ:.1f}$')
ax.legend(loc='upper left')
plt.show()
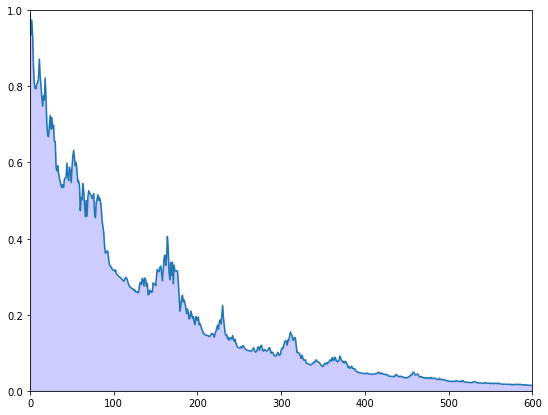
Exercise 25.2
The preceding figure gives some support to the idea that probability mass
converges to
To get a better idea, choose a small
for
Plot
Your figure should show error erratically declining something like this
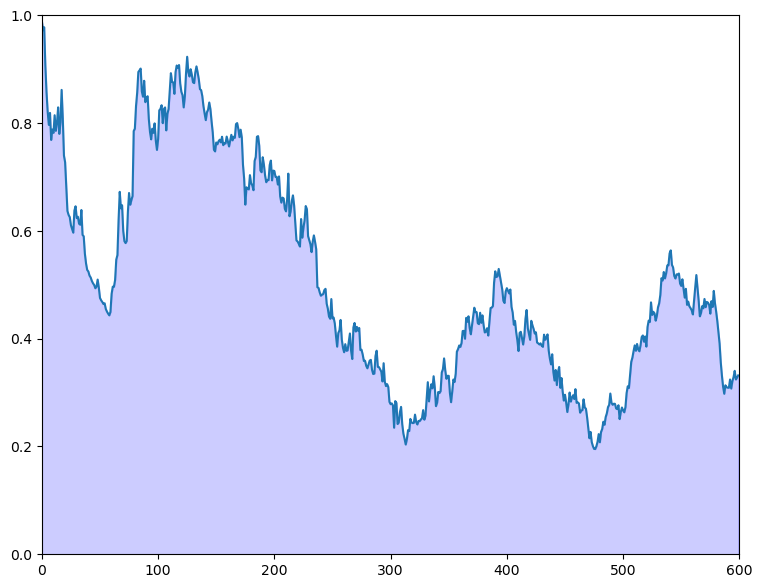
Solution to Exercise 25.2
ϵ = 0.1
θ = 10 # Constant value of state x_t
A, C, G, H = 1, 0, 1, 1
ss = LinearStateSpace(A, C, G, H, mu_0=θ)
x_hat_0, Σ_0 = 8, 1
kalman = Kalman(ss, x_hat_0, Σ_0)
T = 600
z = np.empty(T)
x, y = ss.simulate(T)
y = y.flatten()
for t in range(T):
# Record the current predicted mean and variance and plot their densities
m, v = [float(temp) for temp in (kalman.x_hat.item(), kalman.Sigma.item())]
f = lambda x: norm.pdf(x, loc=m, scale=np.sqrt(v))
integral, error = quad(f, θ - ϵ, θ + ϵ)
z[t] = 1 - integral
kalman.update(y[t])
fig, ax = plt.subplots(figsize=(9, 7))
ax.set_ylim(0, 1)
ax.set_xlim(0, T)
ax.plot(range(T), z)
ax.fill_between(range(T), np.zeros(T), z, color="blue", alpha=0.2)
plt.show()
Exercise 25.3
As discussed above, if the shock sequence
Let’s now compare the prediction
This competitor will use the conditional expectation
The conditional expectation is known to be the optimal prediction method in terms of minimizing mean squared error.
(More precisely, the minimizer of
Thus we are comparing the Kalman filter against a competitor who has more information (in the sense of being able to observe the latent state) and behaves optimally in terms of minimizing squared error.
Our horse race will be assessed in terms of squared error.
In particular, your task is to generate a graph plotting observations of both
For the parameters, set
Set
To initialize the prior density, set
and
Finally, set
You should end up with a figure similar to the following (modulo randomness)
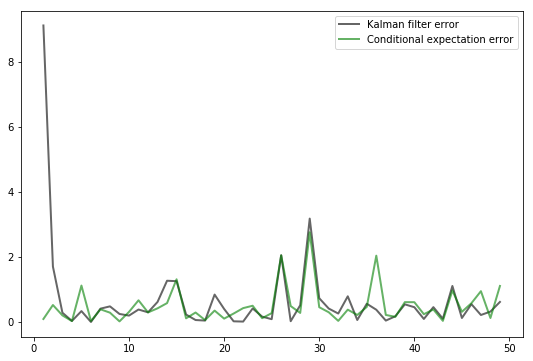
Observe how, after an initial learning period, the Kalman filter performs quite well, even relative to the competitor who predicts optimally with knowledge of the latent state.
Solution to Exercise 25.3
# Define A, C, G, H
G = np.identity(2)
H = np.sqrt(0.5) * np.identity(2)
A = [[0.5, 0.4],
[0.6, 0.3]]
C = np.sqrt(0.3) * np.identity(2)
# Set up state space mode, initial value x_0 set to zero
ss = LinearStateSpace(A, C, G, H, mu_0 = np.zeros(2))
# Define the prior density
Σ = [[0.9, 0.3],
[0.3, 0.9]]
Σ = np.array(Σ)
x_hat = np.array([8, 8])
# Initialize the Kalman filter
kn = Kalman(ss, x_hat, Σ)
# Print eigenvalues of A
print("Eigenvalues of A:")
print(eigvals(A))
# Print stationary Σ
S, K = kn.stationary_values()
print("Stationary prediction error variance:")
print(S)
# Generate the plot
T = 50
x, y = ss.simulate(T)
e1 = np.empty(T-1)
e2 = np.empty(T-1)
for t in range(1, T):
kn.update(y[:,t])
e1[t-1] = np.sum((x[:, t] - kn.x_hat.flatten())**2)
e2[t-1] = np.sum((x[:, t] - A @ x[:, t-1])**2)
fig, ax = plt.subplots(figsize=(9,6))
ax.plot(range(1, T), e1, 'k-', lw=2, alpha=0.6,
label='Kalman filter error')
ax.plot(range(1, T), e2, 'g-', lw=2, alpha=0.6,
label='Conditional expectation error')
ax.legend()
plt.show()
Eigenvalues of A:
[ 0.9+0.j -0.1+0.j]
Stationary prediction error variance:
[[0.40329108 0.1050718 ]
[0.1050718 0.41061709]]
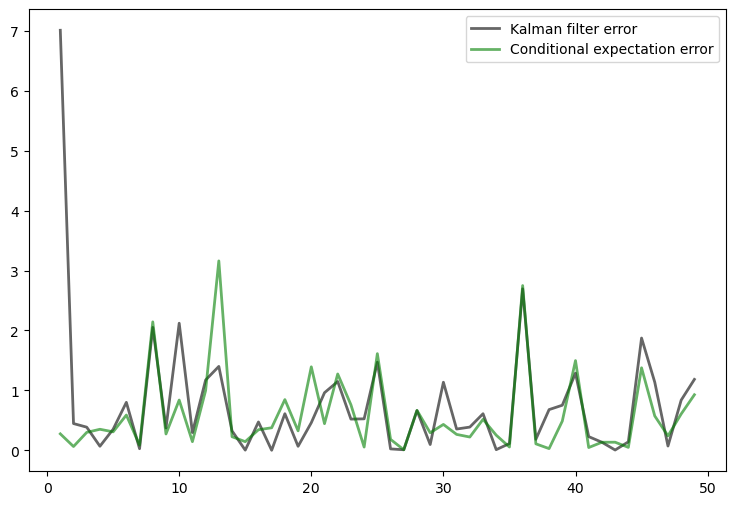
Exercise 25.4
Try varying the coefficient
Observe how the diagonal values in the stationary solution
The interpretation is that more randomness in the law of motion for
- 1
See, for example, page 93 of [Bishop, 2006]. To get from his expressions to the ones used above, you will also need to apply the Woodbury matrix identity.